PACOM is a Java stand alone tool that is able to import, integrate, manage and compare numerous proteomics datasets at the same time, offering a rich set of graphical representation of the most common proteomics data features
We are continuosly trying to improve the software, adding new features and fixing bugs. New versions are automatically uploaded to out server.
Download the latest versions from here or in the release page (https://github.com/smdb21/PACOM/releases)
The updated list of input files supported by PACOM can be found at the corresponding wiki page.
- Java Runtime Environment (JRE) version: 1.8 (Download it from http://www.oracle.com/technetwork/java/javase/downloads/index.html)
- Physical Memory (RAM): As a Java standalone software, you can specify the maximum amount of memory on which it is going to run. It depends on the file (*.bat for Windows and *.sh for Linux and MAC OS) that you use for running it. However, depending on whether the Java Runtime Environment (JRE) you have works on 32 bits or 64 bits (only available for 64 bits OS), you will be able to use more or less amount of memory. For JRE 32 bits version (x86) the maximum amount of memory is only close to 1.4Gb (which is not much). To import datasets into the tool, 1 Gb or 1.4 Gb of RAM memory would be enough, which can be available for 32 bit machines. However, to inspect and compare data, it is strongly recommended to have more memory, such as 16Gb. As an example, 9 experiments containing a total number of 126 fractions (that is, 126 MASCOT search results), will require 8Gb of RAM.
- Recommended Screen Resolution: 1280x700
- Recommended CPU: 2 or more CPU cores (systems with a single core processor will experience a slow performance).
There is no need to install anything. Just download it from the link above, decompress the package (.zip or .tar.gz) and run one of the START_* files (depending on the amount of memory you want to use).
For a more detailed description of these instructions, go to the corresponding wiki page
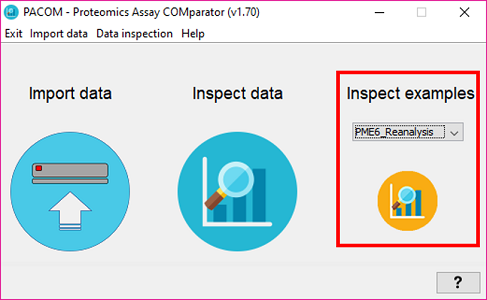
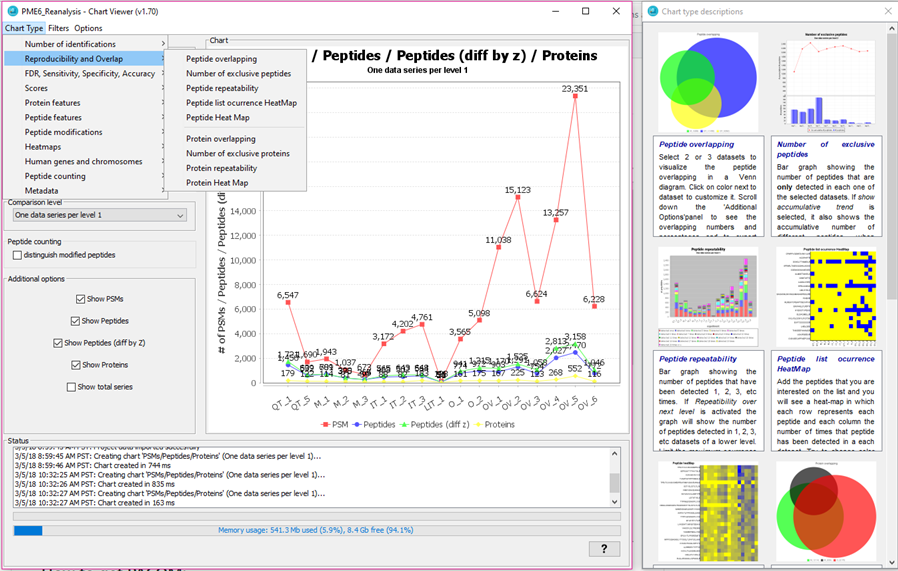
If you want to quickly try PACOM, open it, select one of the example comparison projects (Inspect examples) such as *PME6_Reanalysis" and click on the orange button at the right.
Then, in about 15 seconds you will see the main interface of PACOM for dataset inspection and comparison:
For a complete manual and more detailed information about how to use the tool, go to our wiki page.
Write an email to salvador at scripps.edu. I will try to respond you as soon as possible!
This tool has been enterely designed and implemented by Salvador Martinez-Bartolome firstly as a member of the ProteoRed Bioinformatics Working Group, under the supervision of Juan Pablo Albar, at the Proteomics Laboratory of the National Center for Biotechnology (CNB-CSIC) in Madrid, Spain. Later, the project was continued under the supervision of John R. Yates III at the John Yates laboratory at The Scripps Research Institute, La Jolla, California, USA.
- Miguel Angel Lopez (the best hardware support).
- J. Alberto Medina (software development support and beta tester).
- Gorka Prieto (@akrogp) (PAnalyzer grouping algorithm developer).
- Rosana Navajas (beta tester and manuscript contributor).
- Carmen Gonzalez (beta tester and manuscript contributor).
- Carolina Fernandez Costa (beta tester and manuscript contributor).
- Emilio Salazar-Donate (@emiliosalazardonate) (First developer contributing to this project in its very early phase).
- Juan Pablo Albar (main supporter of this project. All success of this project is dedicated to him).
- John R. Yates III (@proteomicsyates) (supporter of this project in its second phase).
This tool has been developed using the Java MIAPE API which source code can be fount at this git hub web page.